Planar wall potential¶
This module implements the external potential for a collection of planar walls.
Position vector 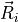 of the wall
of the wall  is given by
is given by
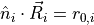 , where
, where 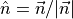 is the normalized outward normal vector to the wall surface.
is the normalized outward normal vector to the wall surface.
 is the distance of
a particle, at position
is the distance of
a particle, at position  , from the wall
, from the wall  .
.
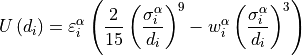
is the interaction energy of a particle of species 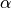 due to
the wall
due to
the wall  . While computing
. While computing 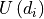 we always
make
we always
make  positive.
positive.
The potential is truncated at a cutoff distance 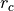 and further
transformed to a
and further
transformed to a 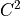 continuous function using
continuous function using halmd.mdsim.forces.pair_trunc.
The parameters  ,
, 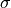 ,
, 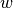 and
and
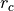 depend on both the wall and the species. For example,
depend on both the wall and the species. For example,
![\varepsilon[i][\alpha]](../../../../_images/math/018dd18d7283212806ae2723d9a6605b99d4bbf7.png) contains
contains 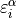 , where
indices
, where
indices  ,
, 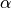 run over the wall and species
respectively.
run over the wall and species
respectively.
-
class
halmd.mdsim.potentials.external.planar_wall(args)¶ Construct the planar wall module.
Parameters: - args (table) – keyword arguments.
- args.offset (table) – positions of the walls
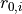 in MD units.
in MD units. - args.surface_normal (number) – outward normal vectors to the wall surfaces
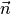 in MD units.
in MD units. - args.epsilon (matrix) – interaction strengths
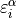 in MD units.
in MD units. - args.sigma (matrix) – interaction ranges
 in MD units.
in MD units. - args.wetting (matrix) – wetting parameters
 in MD units.
in MD units. - args.cutoff (matrix) – cutoff lengths
 in MD units.
in MD units. - args.smoothing (number) – smoothing parameter
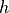 for the
for the 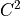 continuous truncation in MD units.
continuous truncation in MD units. - args.memory (string) – select memory location (optional).
- args.label (string) – instance label (optional).
If all elements of a parameter sequence are equal, a single value may be passed instead. In this case,
speciesmust be specified.If the argument
speciesis omitted, it is inferred from the length of the parameter sequences.The supported values for
memoryare “host” and “gpu”. Ifmemoryis not specified, the memory location is selected according to the compute device.-
offset¶ Sequence with the wall position
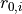 .
.
-
surface_normal¶ Sequence with outward normal vector to the wall surface
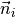 .
.
-
epsilon¶ Sequence with interaction strength
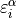 .
.
-
sigma¶ Sequence with interaction range
 .
.
-
wetting¶ Sequence with wetting parameter
 .
.
-
cutoff¶ Sequence with cutoff length
 .
.
-
smoothing¶ Sequence with smoothing parameter
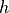 for the
for the 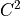 continuous truncation.
continuous truncation.
-
description¶ Name of potential for profiler.
-
memory¶ Device where the particle memory resides.
