Neighbour List¶
This module provides the implementation for a Verlet neighbour list. It stores the neighbours for each particle that are within a certain radius to reduce the computational cost for the force calculation in each time step.
Due to its nature it can only work with finite interaction potentials.
-
class
halmd.mdsim.neighbour(args)¶ Construct neighbour module.
Parameters: - args (table) – keyword arguments
- args.particle – instance, or sequence of two instances, of
halmd.mdsim.particle - args.box – instance of
halmd.mdsim.box - args.r_cut (table) – matrix with elements
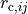
- args.skin (number) – neighbour list skin (default: 0.5)
- args.algorithm (string) – Preferred implementation of the neighbour list (GPU variant only)
- args.occupancy (number) – Desired cell occupancy. Defaults to
halmd.mdsim.defaults.occupancy()(GPU variant only) - args.disable_binning (boolean) – Disable use of binning module and construct neighbour lists from particle positions directly (default: false).
- args.disable_sorting (boolean) – Disable use of Hilbert sorting
halmd.mdsim.sorts.hilbert(default: false). - args.displacement – instance or two instances of
halmd.mdsim.max_displacement(optional) - args.binning – instance or two instances of
halmd.mdsim.binning(optional)
If all elements in
r_cutmatrix are equal, a scalar value may be passed instead.If
displacementorbinningis left unspecified, a default module ofhalmd.mdsim.max_displacementorhalmd.mdsim.binningis constructed. Providing an instance of the respective module allows the reuse of the module (e.g. when different neighbour lists share the first instance ofparticle).For the
hostimplementation of theparticlemodule with binning disabled, Hilbert sorting is disabled also.Specifying
algorithmwill affect the GPU implementation of the neighbour list build when binning is enabled only. The available algorithms arenaiveandshared_mem, where the latter tends to be faster on older GPUs (i.e. ≤ Tesla C1060), but slower on at least GTX 260 and later. Note that theshared_memalgorithm works only when both binning modules have equal number of cells in each spatial direction.-
particle¶ Sequence of the two instances of
halmd.mdsim.particle.
-
displacement¶ Sequence of two instances of
halmd.mdsim.max_displacement.
-
binning¶ Sequence of two instances of
halmd.mdsim.binning. May benilif binning was disabled.
-
cell_occupancy¶ Average cell occupancy. Only available on GPU variant.
-
r_skin¶ “Skin” of the particle. This is an additional distance ratio added to the cutoff radius. Particles within this extended sphere are stored as neighbours.
-
disconnect()¶ Disconnect neighbour module from core and profiler.
