Navigation
- index
- modules |
- next |
- previous |
- HALMD home |
- HALMD v0.2.1 documentation »
- Usage »
HALMD has three ways of accepting program parameters:
Options are described in the command line help:
halmd --help
The group of global options must precede module-specific options, which are introduced by their respective module name.
Many simulation scenarios can be controlled by command line options, there is
no need to setup a separate simulation script. Let us consider a simple fluid
with 20,000 Lennard-Jones particles at density 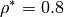 .
Equilibration is done with the Andersen thermostat at a temperature
.
Equilibration is done with the Andersen thermostat at a temperature
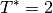 over 10,000 steps
over 10,000 steps
halmd -v \
--force=lennard_jones --integrator=verlet_nvt_andersen \
--timestep 0.005 --time 50 \
box --density 0.8 --particles 20000 \
verlet_nvt_andersen --temperature 2 \
trajectory --every 10000
Many parameters have sensible default values and may be omitted, e.g, the
collision rate of the Andersen thermostat (10), or the cutoff radius of the
potential (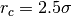 ). The option -v makes the output more
verbose, check that your CUDA device has beed detected properly.
). The option -v makes the output more
verbose, check that your CUDA device has beed detected properly.
The equivalent parameter configuration file would look like this
# file example.rc
force=lennard_jones
integrator=verlet_nvt_andersen
timestep=0.005
time=50
[box]
density=0.8
particles=20000
[verlet_nvt_andersen]
temperature=2
[trajectory]
every=10000
It is passed directly to HALMD, while parameters may be overwritten:
halmd -v -c example.rc --time 10
The trajectory is written every 10,000 steps, i.e., at the beginning and the end of the simulation. The initial configuration of the particles is a fcc lattice. The default output settings yield an HDF5 file with a time stamp in its name, halmd_%Y%m%d_%H%M%S.h5 and a corresponding log file.
We may now continue the simulation at constant energy by resuming from the trajectory file and selecting a different integrator
halmd -v \
--integrator=verlet --timestep 0.001 --time 100 \
trajectory --file halmd_20110715_160545.h5 \
thermodynamics --every 100
This will continue the simulation over 10⁵ steps and write observables like thermodynamic state variables every 100 steps (potential energy, instantaneous “temperature”, pressure, ...
If the HDF5 tools are properly installed, we may have a quick overview of the output file
h5ls halmd_20110715_160920.h5
or look at a specific data set
h5dump -d observables/potential_energy halmd_20110713_161511.h5 | less
For a more advanced inspection and analysis of the HDF5 output files, see Plotting the results. You may try the exemplary script
halmd/examples/plotting/plot_h5md.py halmd_20110713_161511.h5