Velocity Verlet with Nosé–Hoover thermostat¶
This NVT-ensemble integrator implements the Verlet algorithm algorithm
with Nosé–Hoover chain thermostat with a chain length  .
.
For reference and detailed description of the algorithm see the original papers by S. Nosé, W.G. Hoover and Martyna et al.:
- S. Nosé, J. Chem. Phys. 81, 511 (1984)
- W. G. Hoover, Phys. Rev. A 31, 1695 (1985)
- J. Martyna et al., J. Chem. Phys. 97, 2635 (1992)
-
class
halmd.mdsim.integrators.verlet_nvt_hoover(args)¶ Construct velocity-Verlet integrator with Nosé–Hoover chain thermostat.
Parameters: - args (table) – keyword arguments
- args.particle – instance of
halmd.mdsim.particle - args.box – instance of
halmd.mdsim.box - args.timestep (number) – integration time step (defaults to
halmd.mdsim.clock.timestep) - args.temperature (number) – temperature of heat bath
- args.resonance_frequency (number) – coupling frequency of the thermostat
-
set_timestep(timestep)¶ Set integration time step in MD units.
Parameters: timestep (number) – integration timestep This method forwards to
halmd.mdsim.clock.set_timestep(), to ensure that all integrators use an identical time step.
-
timestep¶ Integration time step in MD units.
-
set_temperature(temperature)¶ Set temperature of heat bath.
Parameters: temperature (number) – temperature of heat bath
-
temperature¶ Temperature of heat bath.
-
resonance_frequency¶ Resonance frequency of the Nosé–Hoover thermostat, this is connected to the mass of the thermostat via
 and
and  , where
, where  is
is
 ,
, 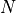 the total number
of (point) particles, and
the total number
of (point) particles, and  the dimension of space.
the dimension of space.
-
set_mass(mass)¶ Set mass of heat bath.
Parameters: mass (table) – Sequence of masses  for the heat bath coupling.
for the heat bath coupling.
-
mass¶ Array of masses
 of heat bath, connected to the coupling
strength of the thermostat.
of heat bath, connected to the coupling
strength of the thermostat.
-
integrate()¶ Calculate first half-step.
By default this function is connected to
halmd.mdsim.core.on_integrate().
-
finalize()¶ Calculate second half-step.
By default this function is connected to
halmd.mdsim.core.on_finalize().
-
position()¶ Return current values of thermostat chain variables (which are generalised positions).
-
velocity()¶ Return current “velocities” of the thermostat chain variables.
-
internal_energy()¶ Return internal energy of thermostat variables divided by particle.nparticle.
-
disconnect()¶ Disconnect integrator from core and profiler.
-
class
writer(args)¶ Write heat bath variables (generalised positions and velocities) and derived quantities (internal energy) to file.
Parameters: - args (table) – keyword arguments
- args.file – instance of file writer
- args.fields (table) – data field names to be written
- args.location (string table) – location within file (optional)
- args.every (number) – sampling interval (optional)
Returns: instance of group writer
The table
fieldsspecifies which data fields are written. It may either be passed as an indexed table, e.g.{"position", "velocity"}, or as a dictionary, e.g.,{xi = "position", v_xi = "velocity"}; the table form is interpreted as{position = "position", ...}. The keys denote the field names in the file and are appended tolocation. Valid values areposition,velocity,internal_energy.The argument
locationspecifies a path in a structured file format like H5MD given as a table of strings. If omitted it defaults to{"observables", "nose_hoover"}.If
everyis not specified or 0, a phase space sample will be written at the start and end of the simulation.-
disconnect()¶ Disconnect Nosé–Hoover writer from observables sampler.
